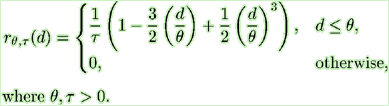
This web page contains Splus functions for generating stationary [not necessarily isotropic] Gaussian random fields over regular grids, and for clipping and visualizing them. There is also a section on computing the Matérn covariance.
The related resources are
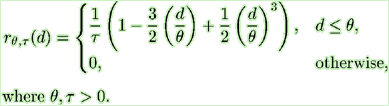
The corresponding Splus code is
|
sphercov <- function (x, y, theta1, theta2, tau) { d <- sqrt (x^2 + y^2) ifelse (d < theta1, (1- 1.5*(d/theta1)+0.5*(d/theta1)^3) / tau, 0) } |
Note that the covariance function must be able to accept vectors for x and y.
Next, copy-and-paste the two following functions into your Splus session.
|
embed <- function (n1, n2, covfun, theta1, theta2, tau) {
xf <- rep (c(rep(1, n2), 0, n2:2), n1) yf <- rep (c(1:n2, 0, rep(1, n2-1)), n1) + n2 * 0:(2*n1*n2 - 1) %/% (2*n2) xb <- rep (c(rep(n1*n2 - n2 + 1, n2), 0, (n1*n2):(n1*n2 - n2 + 2)), n1 - 1) - n2 * 0:(2*(n1 - 1)*n2 - 1) %/% (2*n2) yb <- rep (c(1:n2, 0, rep(1, n2 - 1)), n1 - 1) indexX <- c(xf, rep(0, 2*n2), xb) indexY <- c(yf, rep(0, 2*n2), yb) z <- indexX==0 | indexY==0 x1 <- (indexX-1) %/% n2 + 1 y1 <- (indexX-1) %% n2 + 1 x2 <- (indexY-1) %/% n2 + 1 y2 <- (indexY-1) %% n2 + 1 ifelse (z, 0, covfun (x1-x2, y1-y2, theta1, theta2, tau)) } |
|
genField <- function (n1, n2, covfun, theta1, theta2, tau) { v <- embed(n1, n2, covfun, theta1, theta2, tau) V <- matrix(v, ncol=2*n2, byrow=T) LAMBDA <- fft(V, inverse=T) if (sum(LAMBDA<0)) stop ("\nNegative embedding! \nUse larger grid!") sqlambda <- sqrt(Re(c(t(LAMBDA)))) epsilon <- rnorm(4*n1*n2, 0, sqlambda) + 1i*rnorm(4*n1*n2, 0, sqlambda) EPSILON <- matrix(epsilon, ncol=2*n2, byrow=T) W <- fft(EPSILON, inverse=T) / sqrt(4*n1*n2) w <- Re(c(t(W))) indexZ <- rep(1:n2, n1) + 2*n2 * 0:(n1*n2-1) %/% n2 matrix (w[indexZ], ncol=n2, byrow=T) } |
Once you have the functions cov, embed, and genField, you are ready to generate Gaussian random fields. For example, to generate a 128x128 sample with Spherical(100, 1, 1) correlation, type
| z <- genField (128, 128, sphercov, 100, 1, 1) |
| persp (z) |
| image (z) |
| z <- genField (13, 13, sphercov, 100, 1, 1) |
|
z <- genField (128, 128, sphercov, 100, 1, 1) z <- z[1:13, 1:13] |
We suggest two functions for writing random fields into PPM images. The first function converts continuous (e.g. Gaussian) data into levels of gray.
|
writePPM <- function (field, filename){ if (length(dim(field)) != 2) stop ("\nField must be a matrix!") fmin <- min(c(field)) fmax <- max(c(field)) seg <- (fmax-fmin)/255 greylev <- round((c(field)-fmin) / seg) cat ("P3", dim(field), "255", sep="\n", file=filename, append=F) write (t(cbind(greylev, greylev, greylev)), ncol=15, file=filename, append=T) } |
For example, if z is a realization of the Gaussian random field, as returned by the genField function, you can type
| writePPM (z, "file.ppm") |
| !xv file.ppm & |
The second function is for clipping random fields and coloring the resulting discrete field. To use it, you must first download the file colors.txt and put it into the directory from which you start Splus. Then copy-and-paste the following code:
|
writeClippedPPM <- function (field, thresholds, colors, filename) {
if (length(dim(field)) != 2) stop ("\nField must be a matrix!") n <- length(colors) if (n != length(thresholds)+1) stop("\n#thresholds must be \n #colors less one!") colorDef <- dget("colors.txt") z <- c(field) qz <- matrix (rep(colorDef[colors[n],], length(z)), ncol=3, byrow=T) indz <- z < thresholds[1] len <- sum(indz) qz[indz,] <- c(rep(colorDef[colors[1], 1], len), rep(colorDef[colors[1], 2], len), rep(colorDef[colors[1], 3], len)) if (n>2) for (i in 1:(n-2)) { indz <- z < thresholds[i+1] & z > thresholds[i] len <- sum(indz) qz[indz,] <- c(rep(colorDef[colors[i+1], 1], len), rep(colorDef[colors[i+1], 2], len), rep(colorDef[colors[i+1], 3], len)) } cat ("P3", dim(field), "255", sep="\n", file=filename, append=F) write (t(qz), ncol=15, file=filename, append=T) } |
Now you can type
|
writeClippedPPM (z, c(-0.43, 0.43), c("dodger blue", "white", "grey"), "sky.ppm") |
To view the resulting image, type
| !xv sky.ppm & |
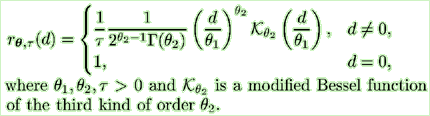
| Splus COMPILE matern.c |
| dyn.load ("matern.o") |
|
matcov <- function (x, y, theta1, theta2, tau) { d <- sqrt(x^2 + y^2) answer <- numeric(length(d)) .C("matern_cov", as.double(d), as.integer(length(d)), as.double(theta1), as.double(theta2), as.double(answer))[[5]]/tau } |
|
z <- genField (128, 128, matcov, 6, 2, 1) persp (z) |